What is Oxford Nanopore Technology (ONT) sequencing?
Image credit: Oxford Nanopore Technologies

Oxford Nanopore Technology developed third generation sequencers that are portable, able to sequence DNA in remote locations and produce ultra-long reads.
- In 2015, the first Oxford Nanopore Technology (ONT) sequencing device was released – an entirely new way to sequence DNA.
- Unlike previous techniques, which were based on DNA replication, ONT doesn’t use any DNA polymerases. Instead, it’s based on a small barrel-shaped protein in a membrane, called a nanopore, and measures changes to electrical current.
- It was used in 2015 during an Ebola outbreak to help with genomic surveillance.
Key terms
DNA
(deoxyribonucleic acid) A molecule that carries the genetic information necessary to build and maintain an organism.
DNA sequencing
The process of determining the order of bases in a section of DNA.
What is Oxford Nanopore Technology?
- Oxford Nanopore Technology (ONT) is a type of third generation sequencing technology.
- Unlike all previous sequencing technologies, ONT doesn’t use any DNA polymerases.
- Instead, it uses a barrel-shaped protein called α-hemolysin – naturally found as a ‘pore’ in a cell membrane, regulating which molecules can enter or leave a cell. This is called a nanopore.
- α-hemolysin has a diameter of 1 nanometre – just big enough to allow a single strand of DNA through.
How does ONT work?
- In ONT, the α-hemolysin nanopore is embedded into an artificial membrane inside a sequencing chamber. When a current is applied to the membrane, the DNA travels through the nanopore.
- As the DNA travels through the nanopore, it obstructs the current flowing across the membrane.
- The four bases of the DNA (A, T, C and G) are of different shape and size, so cause variations in the current.
- These variations are measured by an electronic chip. An algorithm converts the data into a sequence which can then be read.
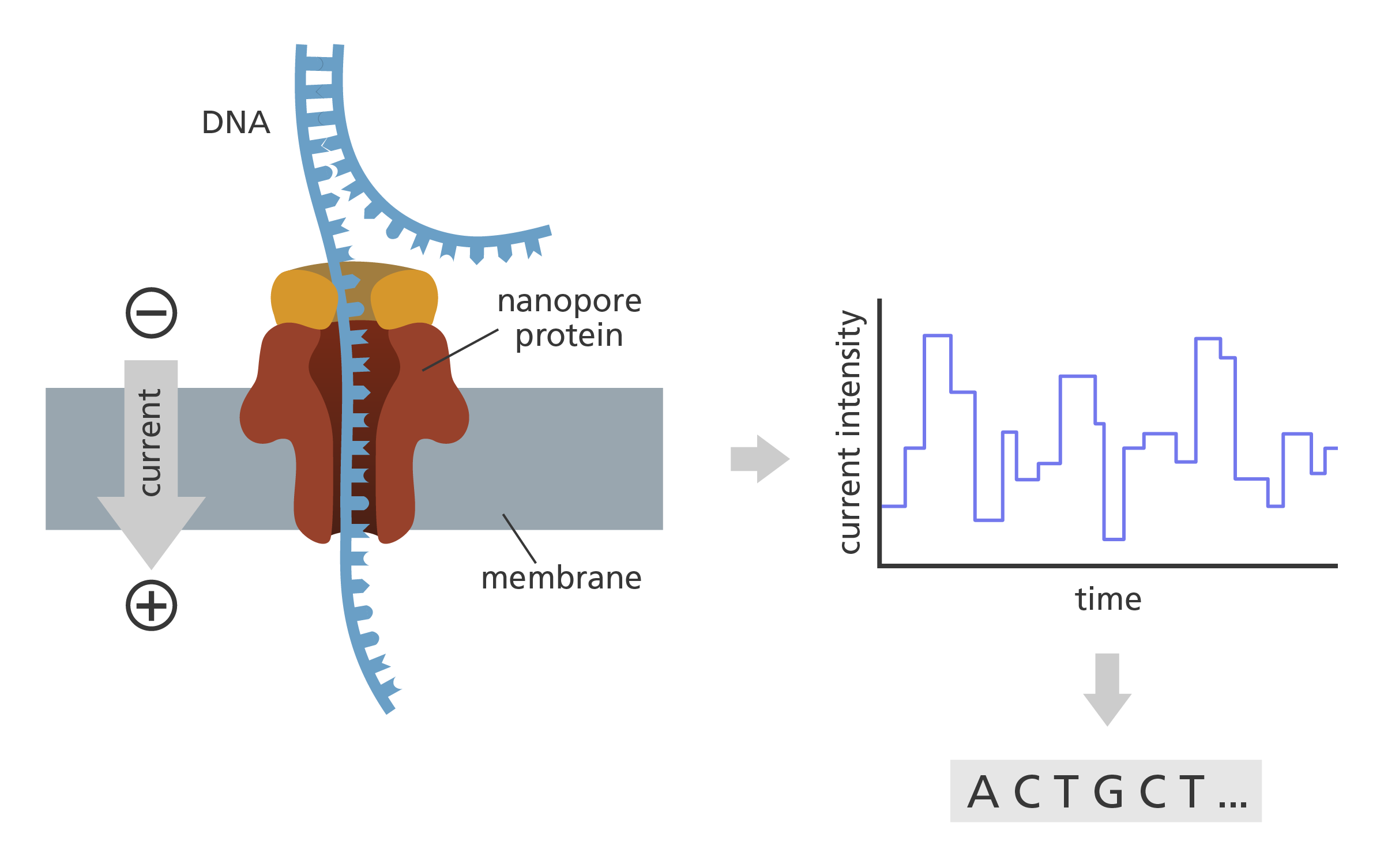
Oxford Nanopore Technology. Nanopore proteins are embedded into an artificial membrane inside the sequencing flow cell. The obstruction of a nanopore by a DNA fragment leads to a change in the current that is measured continuously by an electronics chip integrated within the flow cell. Image credit: Laura Olivares Boldú, Wellcome Connecting Science
What are the benefits of Oxford Nanopore Technology (ONT)?
- Despite having a lower accuracy than other technologies, ONT has several unique advantages.
- It can generate ultra-long reads (up to several millions of base pairs) making it much easier to sequence an entire genome.
- The instrument is very small – depending on the model, between the size of a phone and a microwave. Comparatively, Illumina and PacBio sequencers are closer in size to fridge-freezers. ONT’s small size makes the technology more accessible outside of the traditional laboratory setting, requiring only a laptop to run it.
- For example, the smallest ONT sequencer has been used onboard the International Space Station. It has also been used in low-income countries, including during the Ebola pandemic in 2015 – when it read the genomes of Ebola viruses from 14 patients in just 48 hours.

minION sequencing device being used in Guinea during the 2013-2016 Ebola outbreak
Image credit: Tommy Trenchard / European Mobile Laboratories