What is Sanger Sequencing?

The Sanger sequencing method enabled scientists to read the genetic code for the first time.
- DNA sequencing is the process of determining the exact order of the four bases (A, T, C and G) in a specific section of DNA.
- In the 1970s, Frederick Sanger developed the first sequencing method – known as the ‘chain termination method’ or ‘Sanger sequencing’.
- The principles are still used well into the 2020s, although the technology has become much more efficient, safe and accurate.
Key terms
DNA
(deoxyribonucleic acid) A molecule that carries the genetic information necessary to build and maintain an organism.
Base
(or nucleotide) The basic unit of genetic instructions. DNA is encoded in four chemical bases: adenine (A), thymine (T), cytosine (C ) and guanine (G).
Genome
The complete set of genetic instructions required to build and maintain an organism.
DNA sequencing
The process of determining the order of bases in a section of DNA.
What is Sanger Sequencing?
- In 1977, a breakthrough from Fred Sanger and his team in Cambridge, UK, made it possible to sequence a genome for the first time.
- It is based on the natural process of DNA replication and initially was analysed by eye, using gel-based techniques.
- Sanger and his team were the first to sequence a whole genome using this technique, the 5,000-base-long genome of a virus called PhiX174.
- It became the most widely used form of sequencing for the next few decades and has significantly improved our understanding of the role of genetics and genomics in biology, health and disease.
Sanger sequencing is based on the natural process of DNA replication.
- During natural DNA replication, the double stranded DNA helix is ‘unzipped’ by an enzyme and eventually results in two identical copies of the double-stranded DNA molecule.
- During Sanger sequencing, the DNA helix is broken down by heat or chemicals.
- In early techniques, it resulted in billions of radioactively labelled DNA fragments of varying lengths, which were separated on a gel and analysed by hand.
How does gel-based Sanger sequencing work?
Sanger sequencing has four main components:
- The template: the DNA that will be sequenced.
- A primer: a short, single-stranded stretch of DNA that provides the starting point for sequencing.
- Bases: individual DNA bases (A, T, C and G), called nucleotides.
- DNA polymerase: an enzyme which ‘walks’ along the DNA, adding complementary bases to the sequence.
1.
Step 1: chain termination polymerase chain reaction.
- The DNA double helix is ‘denatured’ (broken down) with heat or chemicals to separate the two strands. These will then act as templates for DNA synthesis.
- The primer, DNA polymerase and bases are added. One or more of these bases is radioactively labelled so that any DNA that is synthesised can be detected.
- The reaction is split into four – one for each base (A, T, C and G). Versions of the bases, known as terminators which stop DNA synthesis, are added to each reaction.
- For example, adding the ‘A’ terminator stops DNA synthesis when an ‘A’ base is added. This results in a mixture of fragments of radioactive DNA of various lengths but all ending in the same base – in this case, the ‘A’ base.
- The same reaction is then also set up for the other three bases, which stops DNA synthesis when the T, C or G base is added:
- a mixture of nucleotides plus a T terminator.
- a mixture of nucleotides plus a C terminator.
- a mixture of nucleotides plus a G terminator.
2.
Step 2: separating the DNA fragments by gel electrophoresis.
- The four different reactions are then loaded on to separate lanes of an acrylamide gel – like a slab of firm, transparent jelly. This is called gel electrophoresis.
- Electrodes are placed at either end of the gel and an electrical current is applied.
- Since DNA is negatively charged, it migrates to the positive end of the gel when the current is applied – opposites attract.
- Small fragments go through the gel quicker than larger fragments.
- This results in the DNA pieces separated according to size and base, ready to be analysed.
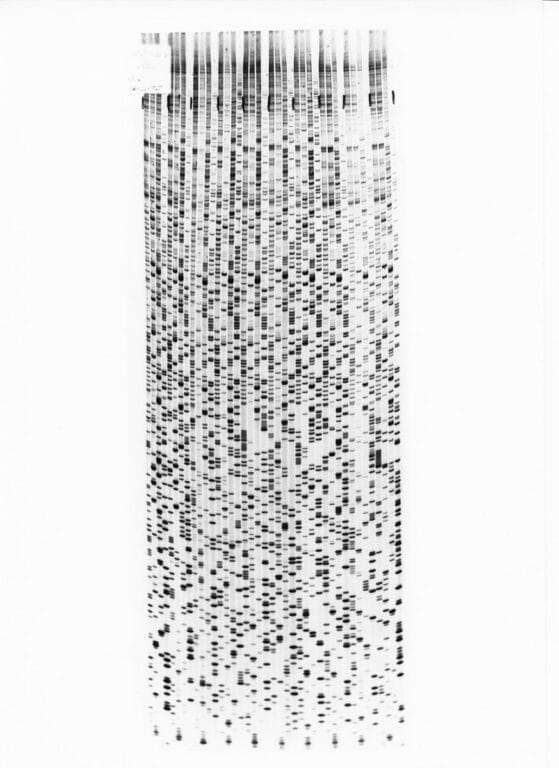 Autoradiogram of a dideoxy sequencing gel, Image credit: Wellcome Genome Campus
Autoradiogram of a dideoxy sequencing gel, Image credit: Wellcome Genome Campus
3.
Step 3: interpreting the data.
- The gel is then visualised by exposing the gel to X-ray film, called an autoradiogram.
- The radioactively labelled DNA will make the film turn black, as in the picture.
- Each band on the film corresponds to where a specific terminator version of one of the bases has been added (A, T, C or G). You can read off the sequence of the DNA from the bottom of the film.
- In the early days of Sanger sequencing, this process was all done by eye and by hand. It could take 12 hours to read the gel.