Genome-wide association studies

Genome-wide association studies compare the genomes of people with and without certain conditions to look for genetic variation that might be responsible.
- Genome-wide association studies scan the genomes of a group of people to find genetic differences that are commonly linked to a particular characteristic, such as the chance of developing certain conditions.
- They’re called ‘genome-wide’ because they look across the entire genome. If a genetic difference is linked to a characteristic or condition, it’s referred to as ‘associated’ with it.
- Studies like these have led to the discovery of thousands of genes that have a role in common diseases, such as type 2 diabetes, Alzheimer’s disease, Parkinson’s disease and inflammatory bowel disease.
What is a genome-wide association study?
- Genome-wide association studies (GWAS) compare many genomes to find common genetic variations that might be associated with a particular characteristic.
- For example, researchers might compare the genome of people with a particular disease to those without the disease.
- These studies often look at the most common type of genetic variation, called single nucleotide polymorphisms (SNPs, pronounced “snips”).
- These SNPs are differences in single DNA bases. They’re a bit like changing a letter in a sentence – depending on the letter, it can change the meaning of the sentence to varying degrees.
- There are up to 5 million SNPs in a person’s genome. Some have no effect. Others change appearance, such as height or eye colour. Others affect health, such as making a person more likely to develop certain conditions and illnesses.
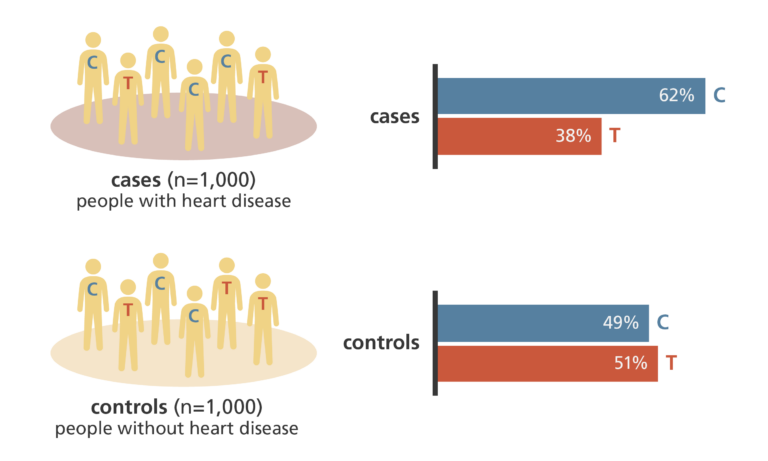
Case-control genome-wide association studies
- A case-control GWAS compares the genomes from two groups of people, such as:
- people with the disease or characteristic being studied (the ‘case’ group)
- people who don’t have the disease or characteristic but are otherwise similar to the first group (the ‘control’ group)
- Hundreds of thousands of SNPs are investigated to see if any are more common in one group than the other.
- This can point towards the genes that are involved in the development of that disease or characteristic.
- It doesn’t necessarily mean the SNP or gene is responsible for the disease or characteristic, but it suggests that this gene or DNA is a good candidate to study further. The next step is to look in detail at that region of the genome in more detail to understand its potential role in the development of the disease or characteristic.
What can we learn from genome-wide association studies?
- Genome-wide association studies let us test a very large number of SNPs at the same time. This is partly thanks to the development of a new piece of technology in 2007, called the ‘SNP chip’.
- This has helped to identify more than 2,000 specific areas of the genome that are associated with diseases, helping us to understand more about how the diseases develop and how we could diagnose or treat them. This includes:
- SNPs that are associated with complex conditions such as type II diabetes, Alzheimer’s disease and Parkinson’s disease. This includes more than 100 variations associated with inflammatory bowel conditions.
- SNPs that influence the way a person will respond to certain medicines.
- insight into the genetics of health conditions, such as finding a strong association between obesity and a gene called FTO.
- It’s currently being used to investigate many complex diseases, such as autoimmune conditions, where the immune system attacks its own body by mistake.