The evolution of genome editing tools
Image credit: Shutterstock

Genome-editing tools have evolved since the 1980s, enabling scientists to make increasingly precise edits to a genome.
- Genome editing can be used to change the characteristics of a cell or organism, by making specific changes to its DNA.
- The way these edits are delivered to the genome is called the editing system.
- Genome-editing systems have evolved since the 1980s, enabling scientists to make increasingly precise edits – such as adding or removing chunks of DNA or making specific alterations to the sequence.
Different genome editing systems
- There are several different types of genome-editing systems in use today.
- Typically, they involve an enzyme called an ‘engineered nuclease’, which helps to bind to, and sometimes cut, the DNA.
- They differ in how they recognise the specific region of DNA – using either a sequence of RNA or a protein to find and bind to the DNA target.
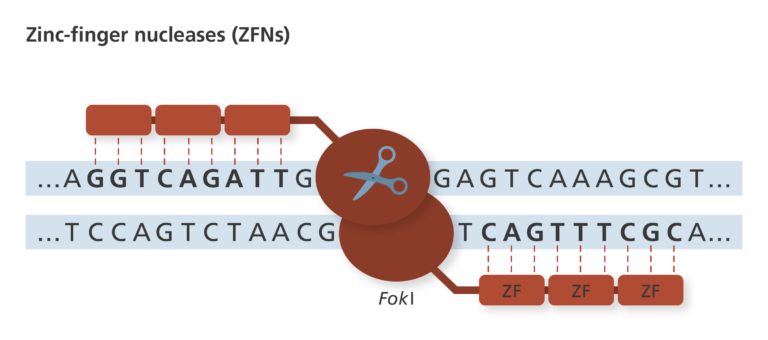
1980s – 2010s: Zinc-finger nucleases (ZFNs)
- Discovered in the 1980s, ZFNs were a common editing tool for several decades.
- ZFNs are made of two parts: a zinc-finger protein, which recognises and binds to the DNA, and the nuclease, which cuts the DNA.
- The zinc-finger protein looks for three DNA bases. Custom ZFNs can be made by combining zinc-finger proteins to recognise specific sequences, in multiples of three bases.
- The nuclease part is normally a FokI nuclease, which cuts the DNA.
- ZFNs work in pairs, with two zinc-finger proteins recognising each strand of DNA and two FokI molecules coming together in the middle to make a cut in the DNA.
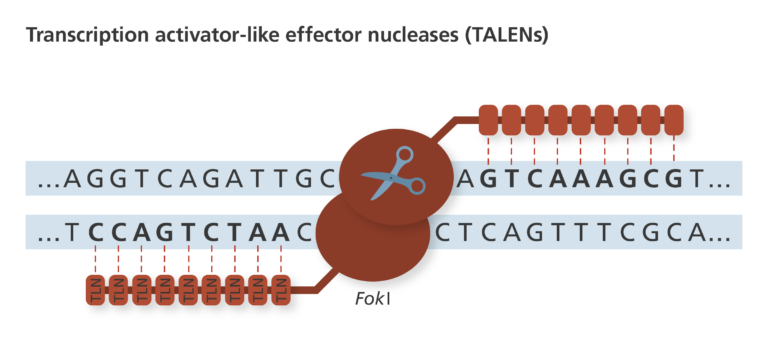
2011 - present: Transcription activator-like effector nucleases (TALENs)
- Discovered in 2011, TALENs continue to be a popular genome-editing system.
- TALENs work in a similar way to ZFNs, except they recognise the DNA sequence using transcription activator-like effector (TALE) domains.
- TALENs can recognise and bind to single bases within a DNA sequence.
- This makes TALENs easier to engineer to recognise specific DNA sequences, because they can be combined in more specific patterns.
- Although in recent years they have become less popular due to the rise in CRISPR-based systems, TALENs still retain some advantages, such as their high specificity and low off-target effects.
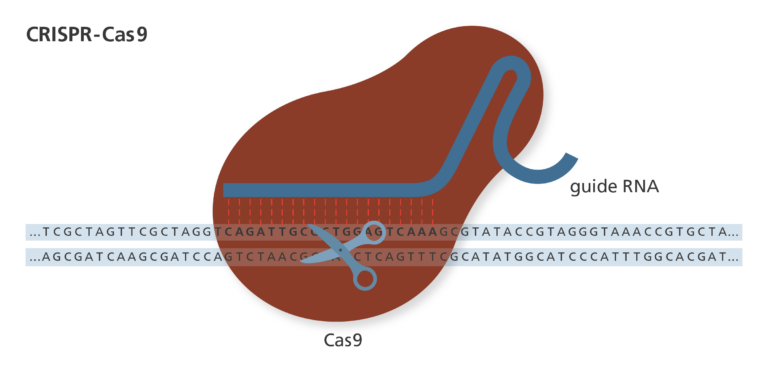
2012 - present: CRISPR-Cas9 (pronounced “crisper-cass-nine”)
- Discovered in 2012, CRISPR-Cas9 is an increasingly popular tool for genome-editing. It’s adapted from a natural genome editing system used by bacterial cells to destroy attacking viruses.
- CRISPR-Cas9 is made of two parts: CRISPR, which uses RNA to target and bind to the DNA, and the nuclease Cas9, which cuts the DNA.
- CRISPR stands for ‘clustered regularly interspaced short palindromic repeats’. Cas9 stands for CRISPR-associated protein 9.
- The CRISPR RNA section can be easily engineered to target nearly any region of DNA. This makes CRISPR-Cas9 cheap and efficient, and it is currently the most used genome editing technique.
- Researchers continue to refine the CRISPR system, including combining it with enzymes other than nucleases, so that it can directly manipulate DNA bases without cutting the DNA. This can result in more reliable genome editing.