What is DNA profiling?
Image credit: Shutterstock

DNA profiling is used to identify an individual from a sample of DNA by looking at unique patterns in the DNA sequence.
- On average, about 99.9% of the DNA between two humans is the same. The remaining 0.1% might not sound like much – but that’s around 3 million bases that can differ between two people.
- DNA profiling looks at these differences to produce a pattern that’s unique to an individual. This can be used for solving crime and linking biological relatives.
- Early techniques were called DNA fingerprinting. Just like your actual fingerprint, your DNA fingerprint is something you are born with, and it is unique to you.
What is a DNA fingerprint?
- Each human shares, on average, about 99.9% of their DNA with each other. The remaining 0.1% represents around 3 million bases and is what makes us unique.
- These differences can be compared and used to help distinguish different people from each other.
- One way of doing this is by comparing short sequences of repetitive DNA that show greater variation from one person to the next than other parts of the genome.
- Early techniques produced this pattern by simultaneously comparing sequences of up to 100 bases long, called ‘minisatellites’. This produces a pattern unique to an individual, known as a DNA fingerprint.
- Modern techniques compare even shorter sequences, called ‘microsatellites’.
- Except for identical twins, the probability of two people having the same DNA fingerprint is very small.
- Despite the similar name, a DNA fingerprint has nothing to do with the fingerprint on the tips of the fingers. A DNA fingerprint is the same for every cell in the body and can’t be altered by any known treatment.
How were early DNA fingerprints produced?
- DNA fingerprinting was invented in 1984 by Sir Alec Jeffreys after he realised you could detect variations in human DNA, in the form of minisatellites.
- Early DNA fingerprinting used ‘minisatellites’ – stretches of DNA which are slightly longer than the ‘microsatellites’ used today.
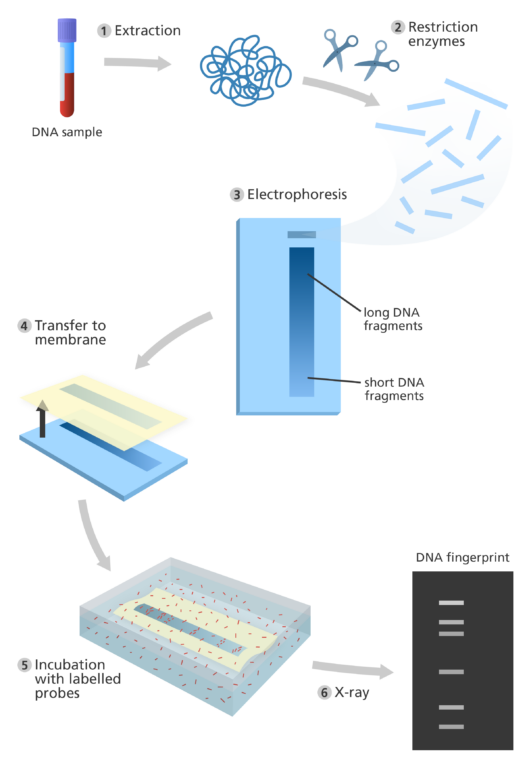
The steps involved in early DNA fingerprinting:
- The first step of DNA fingerprinting was to extract DNA from a sample of human material, usually blood.
- Restriction enzymes – which act like molecular ‘scissors’ – were used to cut the DNA. This resulted in thousands of pieces of DNA with a variety of different lengths.
- These pieces of DNA were then separated according to size by a process called gel electrophoresis:
- The DNA was loaded into wells at one end of a porous gel, which acted a bit like a sieve. An electric current was applied which pulled the negatively charged DNA through the gel.
- The shorter pieces of DNA moved through the gel easiest and fastest. It is more difficult for the longer pieces of DNA to move through the gel, so they travelled slower.
- As a result, by the time the electric current was switched off, the DNA pieces had been separated in order of size. The smallest DNA molecules were furthest away from where the original sample was loaded onto the gel.
- Once the DNA had been separated, the pieces of DNA were transferred from the gel onto a robust piece of nylon. This is called ‘blotting’. The DNA was then ‘unzipped’ to produce single strands of DNA.
- Next the nylon membrane was incubated with radioactive probes – small fragments of microsatellite DNA. They attach to the minisatellite DNA from the genome that they are complementary to.
- The minisatellites that the probes have attached to were then visualised by exposing the nylon membrane to X-ray film. This resulted in a pattern of more than 30 dark bands appearing on the film where the labelled DNA was.
- This pattern was the DNA fingerprint. To compare two or more different DNA fingerprints, the different DNA samples were run side-by-side on the same electrophoresis gel.
Modern DNA profiling
- Modern-day DNA profiling is also called short tandem repeat (STR) analysis.
- It relies on microsatellites, also known as short tandem repeats (STRs). These are the shorter relatives of minisatellites, usually two to five base pairs long.
- Like minisatellites they are repeated many times throughout the human genome, for example ‘TATATATATATA’.
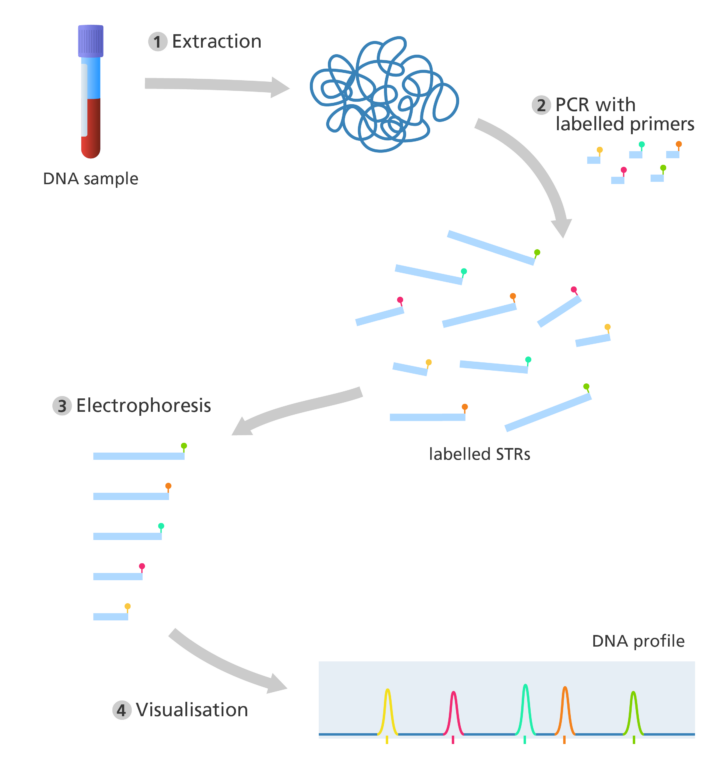
How is a DNA profile produced today?
- DNA is extracted from a biological sample – only a tiny amount of DNA is needed to produce an accurate result. The DNA can be extracted from a wide range of biological samples, including blood, saliva and hair – even if the sample is partially degraded.
- Modern techniques don’t use restriction enzymes to cut the DNA. Instead, they use the polymerase chain reaction (PCR) to produce many copies of specific STR sequences.
- Small pieces of DNA, called ‘primers’, bind to complementary sequences of the DNA either side of the STR of interest. This marks the starting point for the copying of DNA.
- The primers for each STR is labelled with a specific coloured fluorescent tag. This makes it easier to identify and record the STR sequences after PCR.
- Once enough copies of the sequence have been produced, electrophoresis is used to separate the fragments according to size.
- As the DNA fragments separate, they pass by a laser, which causes the fluorescent tags to glow with a specific colour. The output is displayed as a series of coloured peaks (as shown in the image below) highlighting the colour and length of each STR sequence.
How accurate is DNA profiling?
- Only one person in every 10 million million (10,000,000,000,000) will have a particular STR profile.
- It’s extremely unlikely to share the same profile with someone else – except for identical twins.