What was sequencing like before the Human Genome Project?
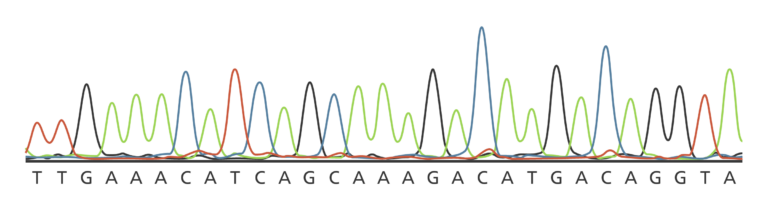
The Sanger sequencing method, developed in 1977, enabled scientists to read the genetic code for the first time. Over the next two decades, the technology evolved until it was fit for use by the Human Genome Project.
Key terms
Base
(or nucleotide) The basic unit of genetic instructions. DNA is encoded in four chemical bases: adenine (A), thymine (T), cytosine (C ) and guanine (G).
Genome
The complete set of genetic instructions required to build and maintain an organism.
DNA sequencing
The process of determining the order of bases in a section of DNA.
Before the Human Genome Project, sequencing techniques were expensive and laborious – albeit pioneering. Their evolution to automated, efficient technologies made it possible to sequence most of the human genome in 13 years.
1977: the dawn of DNA sequencing
DNA sequencing began in 1977 with the development of the ‘Chain Termination Method’ and gel-based analysis. Since it was pioneered by Fred Sanger, it’s also known as the Sanger Method.
Fred Sanger’s DNA sequencing method was based on the natural process of DNA replication, where new strands of DNA are made using an existing strand as a template. By incorporating radioactively labelled DNA bases and ‘terminator’ bases, it can result in billions of copies of the DNA sequence of varying lengths.
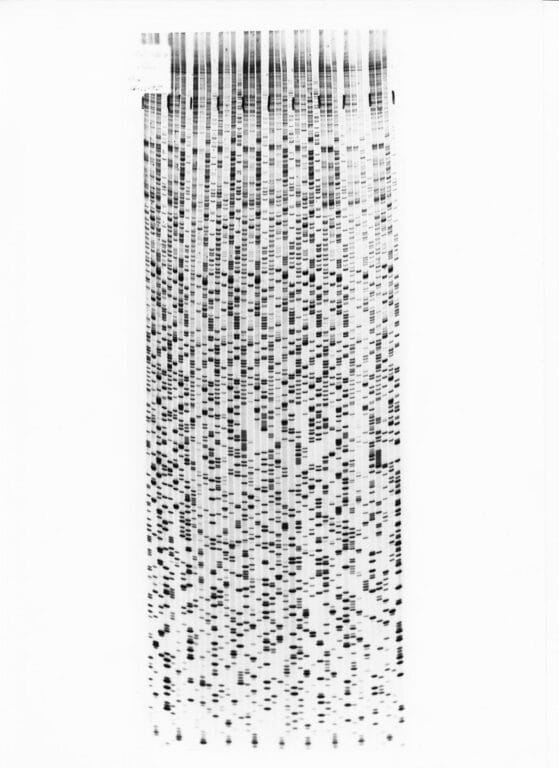
The DNA fragments are separated according to size using a gel-based technique called electrophoresis. It’s then visualised with an x-ray of the gel, called an autoradiogram. You can read more about the process here.
Compared to today’s sequencing techniques, it was a long process. The electrophoresis and the development of the audiogram both took about 12 hours – and it took many hours to read the sequence by eye and hand, before manually entering the data into the computer.
But before Sanger Sequencing, no technique existed to read the genetic code. Sanger and his team would go on to use it to sequence many genomes, including the first whole DNA-based genome using this method – the 5,000-base-long genome of a virus called phiZ174.
The method became the foundation for the DNA sequencing technologies still in use today. Over the next few years, it would become safer, faster and more accurate.
No methods existed to read the genetic code, even in the simplest of genomes, so Fred Sanger was a real pioneer.
1980s: safer, improved, automated sequencing
Sequencing needed to be scaled up – especially if we were going to sequence the DNA of larger organisms, such as humans.
The methods of the 1970s built vital foundations for DNA sequencing – but were laborious, prone to mistakes and used potentially dangerous radioactivity. To sequence the DNA of larger organisms – such as humans – we’d need a safe, scalable, efficient technique.
In the 1980s, a new automated method replaced radioactive bases with coloured dyes: A = Green, C = Blue, G = Yellow and T = Red. DNA fragments were still separated by size on a gel, but the colours meant they didn’t have to be loaded into separate lanes. A machine read and recorded the sequence, storing the data for later.
Automated sequencing machines became commercially available in the late 1980s, making it much quicker, safer and easier to sequence DNA.
However, each base had to be double-checked by eye – and using them to sequence a human genome would still have taken 600 years.

Automatic DNA sequencing machines became commercially available in the late 1980s, allowing scientists to carry out DNA sequencing more safely and efficiently.
1990s: scaling up for the Human Genome Project with capillary sequencing
Sequencing evolved further until we reached capillary sequencing – a faster, easier way to analyse genetic information than gel-based techniques.
Sequencing techniques continued to evolve, leading to capillary sequencing – a more effective method that could process 4-times as many samples as the gel-based techniques. It was this technique that was used throughout the Human Genome Project.
In capillary sequencing, DNA fragments are separated by size through a long, thin, acrylic-fibre capillary. A fluorescence-detecting laser, built into the machine, then shoots through the fibre to make the differently coloured bases fluoresce.
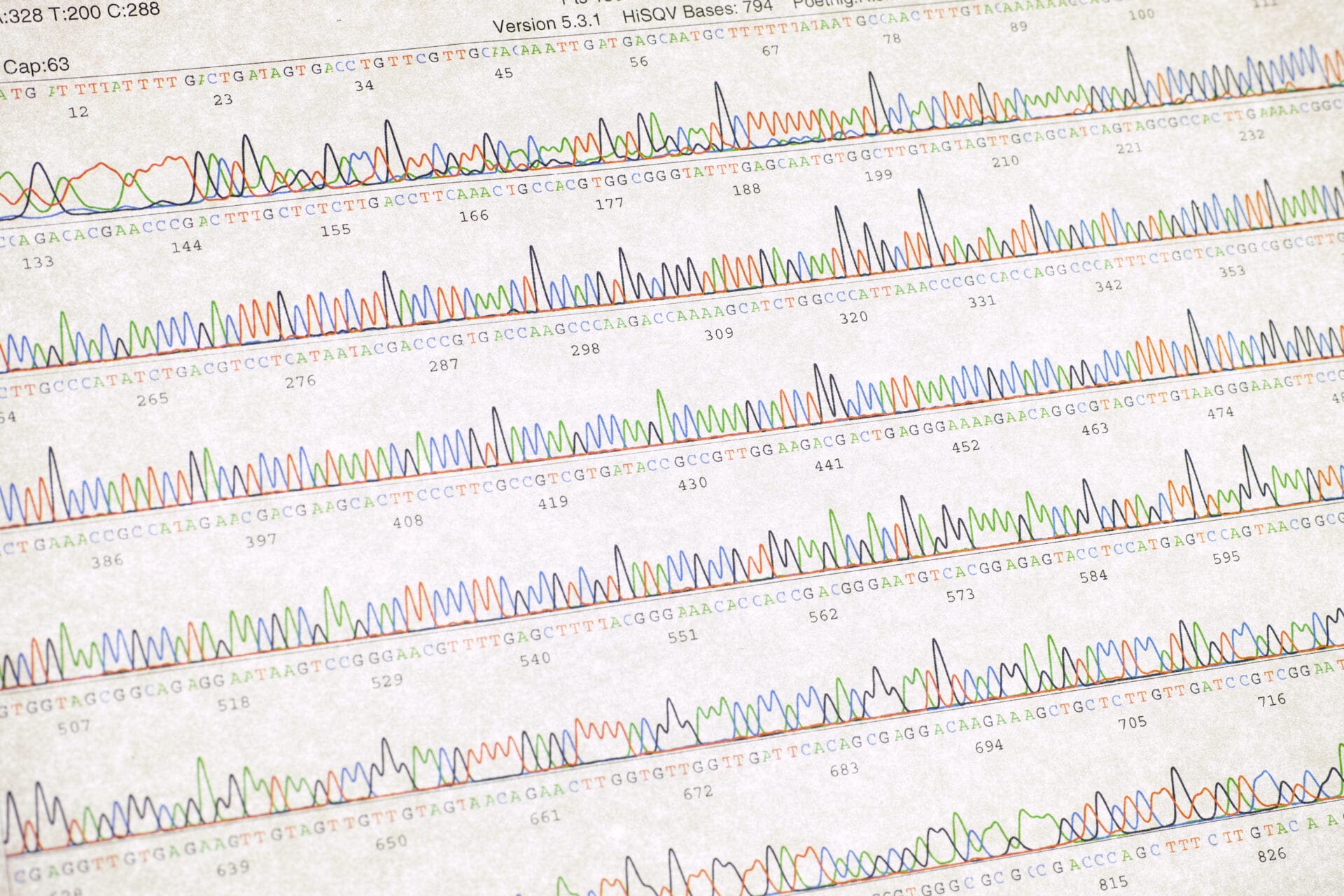
The colour of the fluorescence is detected by a camera and then displayed as a graph of different coloured peaks (see the image below for an example). You can read more about the process here.
Capillary sequencing was much more efficient than gel-based techniques. The small diameter of the capillary allowed for very high electric fields to rapidly separate the DNA fragments by size. And since it was automated, one person could monitor dozens of instruments at once, without having to check each lane of the gel as before.
This helped remove a huge bottleneck from the sequencing process – reducing the time to sequence a whole genome by 50-times.
These important developments made it possible to sequence the human genome – and the technique was used throughout the Human Genome Project.
