Personal genomics: the future of healthcare?

Personal genomics sequences and analyses an individual’s genome, before giving them detailed information about their genomic makeup.
- Every person’s genome is unique and holds clues to their ancestry and personal risk of different conditions.
- With the development of DNA sequencing techniques, it’s becoming practical and affordable for individuals to have their genome sequenced. This is called personal genomics.
What is personal genomics?
The Human Genome Project sequenced DNA pooled from a range of individuals, to create an average or ‘reference’ genome. However, every genome is unique. With the development of DNA sequencing technologies, it’s now increasingly practical and affordable for individuals to choose to get their genomes sequenced.
This is called personal genomics. The first people to have their ‘personal’ genomes sequenced were Craig Venter, founder of Celera Genomics, and James Watson, co-discoverer of the DNA double helix.
Steve Jobs, co-founder of Apple Inc., was also one of the first 20 people in the world to have their DNA sequenced, for which he paid $100,000. He also had the DNA of his cancer sequenced, in the hope it would provide information about more appropriate treatments for him and for other people with the same cancer.
Steve Jobs, co-founder of Apple Inc, was one of the first 20 people in the world to have his DNA sequenced, for which he paid $100,000.
Why carry out personal genomics?
Although the ability to choose to access your own genetic information is still a relatively young phenomenon, it is thought that one day it could form a key part of our everyday healthcare. Personal genomics could allow us to optimise our health on a whole different level to improving our diet and doing more exercise.
By sequencing individual genomes, researchers can uncover large amounts of information concerning all aspects of that individual’s physiology, from their susceptibility to certain diseases to the way they respond to specific drugs.
Personalised medicine
One area that personal genomics is particularly useful in is pharmacogenomics. The genetic information from an individual can, in some cases, be used to select the most appropriate drug to prescribe to a patient. This helps doctors to ensure that the drug has the maximum effect while minimising any potential side effects.

Diagnosing genetic conditions and predicting risk
Personal genomics can also be used to predict or confirm a genetic disease.
By looking at an individual’s genome, it’s possible to identify genetic variants that may increase the likelihood of an individual having a genetic disease later on in life. For example, it can be used to tell a person if they carry the breast cancer-causing variant of the BRCA1 gene and, if so, how much it increases their risk of developing breast cancer.
This gives the individual the option of taking preventative measures – for example, they may choose to have their breast tissue removed if the risk is very high (a mastectomy).
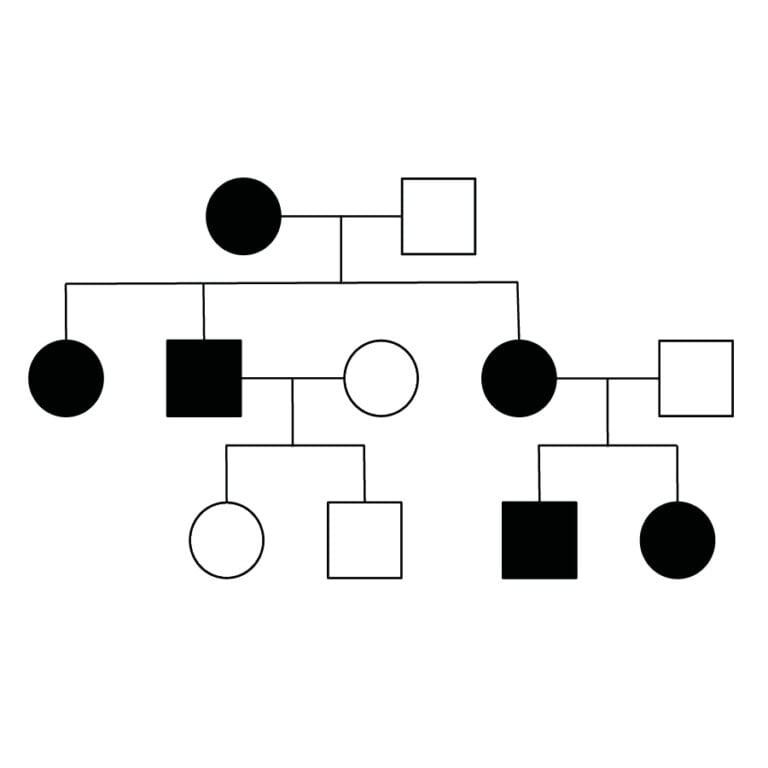
Predicting inheritance
Personal genomics can also be used to predict the risk of passing certain genetic conditions to the next generation.
For example, it is possible to be a ‘carrier’ of recessive genetic conditions like cystic fibrosis, which means that an individual has one of the two genes for the condition but does not exhibit symptoms. However, if their partner is also a carrier the chance of their child having the full condition is dramatically increased.
Screening embryos
By knowing the risk of passing on a genetic condition to their child, some people may decide to investigate other ways of having a baby, such as in vitro fertilisation (IVF). This can enable embryos to be screened for genetic conditions prior to implantation in the womb.
Direct to consumer testing and research projects
The commercialisation of personal genome sequencing is set to grow, and, in future, it could become a routine part of clinical practice.
This is important as the more we know about our individual genes and disease risk, the more easily we will be able to make informed decisions to lessen the likelihood of developing these diseases or delay their onset. However, it’s not always possible to influence how certain conditions will develop over time.
Ultimately, we can only learn more about genetic risks by comparing many genomes in research studies. Some people with personal genome sequences may choose to participate in research projects and allow scientists to study their genomic data to help advance knowledge in the field.
Sharing data is critical to scientific progress – but has been hampered by traditional research practices. Announced in 2005, the Personal Genome Project is a large, long-term, international study looking to create public genome, health and characteristics data, inviting participants to publicly share their personal data for “the greater good”.
With this approach they hope to dramatically speed up research into personal genomics and go some way to fulfilling the curiosity many of us have about our genomes.
All the volunteers provide a cheek swab and a blood or skin sample, from which their DNA is isolated. They also provide information about their characteristics, including medical records and various measurements such as height and weight. This allows the research to examine the relationship between phenotype, our genes and our environment.
Non-anonymous participation and informed consent
After sequencing and analysis, each of the volunteer’s genetic results are sent back to them and, a month later, posted online for all to see.
Although no names or addresses are posted alongside the results, participation in the project had to be ‘non-anonymous’. This is because one of the most identifiable pieces of personal information is our DNA sequence. Even if a name or photograph is not provided, it may, in some cases, be possible to identify someone from the information in their DNA sequence.
This means that the privacy of all Personal Genome Project volunteers cannot be guaranteed. To show that they understand this, all volunteers take several short online tests. By passing the tests, the volunteers show that they can provide ‘informed’ or ‘open’ consent for their genetic material to be publicised without the promise of privacy. The tests also make sure they understand the possibility of finding out about diseases they may be at risk of developing later in life.
The Personal Genome Project is the first project of this kind based on informed consent. This aspect of the project has been considered controversial by some – but by ensuring fully informed consent is an integral part of the project, all participants are sufficiently informed of the risks before signing up.
Open access
The Personal Genome Project is committed to the sharing of data for the advancement of science. By making all the data collected, publicly and freely available to all, they hope that it will reach scientists, drug companies and research institutes across the world.
The Personal Genome Project is effectively building a ‘treasure trove’ of genetic information. This paired with information about health and physical characteristics could dramatically accelerate scientific research. It could help scientists learn more about how genetic and environmental factors interact to cause disease and drive new treatments for diseases such as diabetes and cancer.
The UK arm of the Personal Genome Project was launched in 2013 and is based at University College, London.
