Post-sequencing: sequence comparisons
Image credit: Greg Moss / Wellcome Sanger Institute
How do you find out the significance of a genome after sequencing?
This is part 4 in our series looking at what happens after DNA is sequenced. You might want to start with part 1, looking at the first step of quality control.
- We’ve sequenced the genome, put it back together and identified the genes.
- Now we need to find out what this genome can tell us and how it compares to other genomes.
- This step is our comparison step.
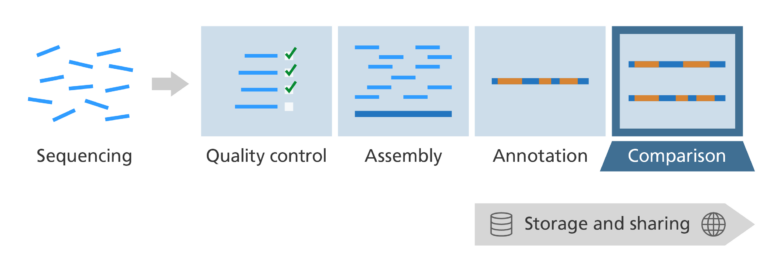
What’s the challenge?
- We have identified genes and other areas of interest, but we don’t yet know their overall significance.
- How can we use the information to answer biological questions?
What do we need to do?
- We need to compare the genome we’ve sequenced with other genomes to identify similarities and differences.
- The outcome of this genome comparison will depend on what question is being asked:
De novo sequencing
- As outlined in part 2 of this series, de novo sequencing is when the genome of an organism is sequenced for the first time.
- Comparing genomes after de novo sequencing is based around finding differences and similarities between the genomes of other organisms.
- Scientists compare the DNA sequences to determine how closely related two species are.
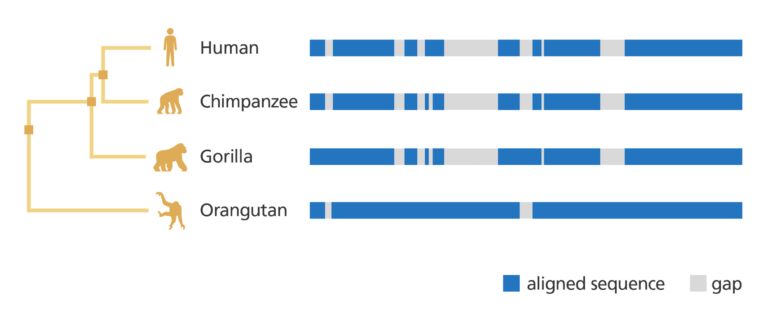
This may provide an insight into the organism and how it ‘works’.
- For example, the genome sequence data of an animal, or model organism, can be annotated and then compared to the annotated sequence of a human.
- By comparing genomes, it’s possible to identify any similar genes. The mouse genome, for example, is very similar to the human genome.
- This information can then be used to investigate similarities in the phenotypes of the mouse and human. For example, a genetic variant is linked to deafness in the mouse – but is this the case in the human as well?
- Organisms can also be created with specific genetic mutations to investigate the function of a particular gene. For example, this gene is linked to ear development – but what is the effect when that gene is not functioning or is absent altogether?
Genome sequencing has also been used to help tackle drug resistance in hospitals.
- For example, the bacterium that causes tuberculosis, Mycobacterium tuberculosis, has become a global health problem because some strains have developed resistance to many antibiotics (‘multidrug resistant’ tuberculosis) making them almost impossible to treat.
- By identifying new resistant strains of the bacterium and sequencing their DNA it is possible to find out more about how drug resistance came about.
- Scientists have found that Mycobacterium tuberculosis evolve resistance in several gradual stages.
- By identifying mutations in the genome that are linked to antibiotic resistance it may be possible to monitor strains that are about to evolve to the next stage of resistance.
- Drugs can then be developed to try and interfere with this evolution before it happens.
Resequencing
- As outlined in part 2 of this series, resequencing is when the genome being sequenced is known to be from a species that has been sequenced before and so a reference genome is available.
- Genetic variants found during annotation are assessed for significance. Do they mean something or are they just there as ‘passengers’ – physically linked to the variant of interest but not contributory to its biological effect in any way?
Example of resequencing
- Any two humans are, genetically, around 99.9% similar. The tiny percentage of genetic material that varies among people can provide clues to key biological questions. For example, is there a variant sitting within a gene that is a known cancer-causing mutation? Has this variant been seen before in someone else? If so, do the disease characteristics of those people overlap?
- The 1000 Genomes Project, which launched in 2008, aimed to produce a catalogue of these differences taken from sequencing the genomes of over 1,000 anonymous people from 26 populations around the world. They hoped that by studying the similarities and differences between each genome they would be able to explain individual differences in susceptibility to disease, response to drugs or reaction to environmental factors.
- The UK10K, launched by the Wellcome Trust in 2010, aimed to analyse the genomes of 4,000 healthy people with those of 6,000 people currently living with a disease of suspected genetic cause, such as severe obesity. By comparing genome sequences, the study has identified genetic variants associated with health and disease, including cholesterol levels and bone health. It is hoped that the information generated by the project will transform our understanding of human genetic variation.
- During these projects each sequenced genome is assembled and annotated using the original human genome reference sequence as a template.
- Each of these genomes is then compared against each other and against the reference sequence to find key differences and similarities that might identify genetic variants associated with specific characteristics or disease.
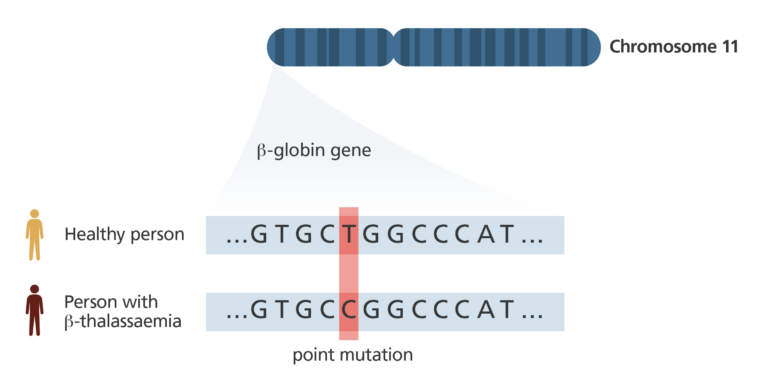
Image credit: Laura Olivares Boldú / Wellcome Connecting Science.