Using genomics to understand malaria

Sequencing the genome of the malaria parasite has revealed interesting clues as to how it is able to evade the human immune system for long enough to cause disease.
This is part 1 in our series looking at how scientists are using genomics to tackle malaria.
Read about how genomics is bringing us closer to finding new weapons to treat malaria effectively and to prevent the disease with new vaccines - giving hope that we might defeat the disease in the future.
- Faster, cheaper genome sequencing has provided a platform to delve deeper into the biological functions of lots of organisms – including Plasmodium falciparum, a dangerous malaria-causing parasite.
- Since the parasite’s genome was sequenced in 2002, scientists have identified the genes that help it to function inside a person’s body to cause malaria.
- This includes the discovery of a family of genes called var, which has provided clues about how the parasite thrives inside the body to drive the disease.
Key terms
Gene
A section of DNA within a genome that carries a specific set of information - often the information needed to make a protein.
Genome
The complete set of genetic instructions required to build and maintain an organism.
Parasite
An organism that lives in or on another organism (the host) and benefits at the expense of their host.
Host
An organism that houses another, smaller organism. This can be positive, negative or neutral to the host organism.
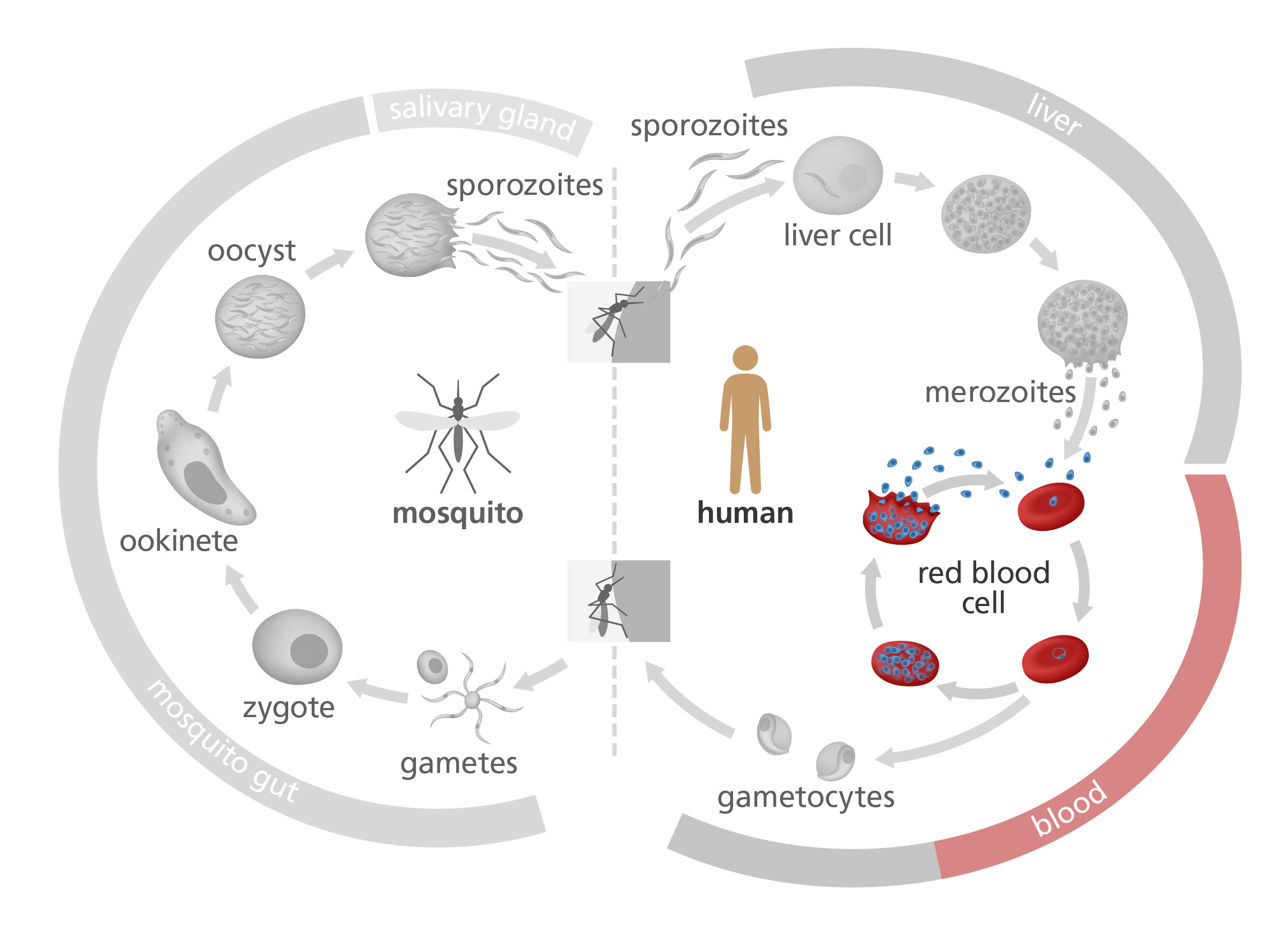
How does Plasmodium cause malaria?
Several Plasmodium parasites cause malaria, but the species that causes the deadliest form of the disease is Plasmodium falciparum. For this reason, it’s usually the main focus of malaria research.
Take a look at the image below, illustrating the life cycle of Plasmodium falciparum. After it’s injected by a mosquito into a human body, it attaches to and infects red blood cells – providing a safe space to hide undetected from the host immune system.
Once the parasite has multiplied a number of times inside the host’s red blood cells, the blood cells bulge and eventually burst, releasing the parasite into the blood.
Scientists are particularly interested in this ‘blood’ stage of the Plasmodium life cycle. This is partly because it’s the stage that causes malaria’s worst symptoms – like fever, muscle aches and iron deficiency – and partly because understanding how the parasite manages to avoid detection by the immune system might lead to new treatments for the disease.
How does Plasmodium avoid the immune system?
Using genomics, scientists have discovered a group of proteins called ‘P. falciparum Erythrocyte Membrane Protein-1s’ – or PfEMP1s for short.
These are large, sticky proteins that appear on the surface of a red blood cell towards the end of the blood stage of the life cycle, helping the parasite interact with its host, from within the safety of the red blood cell.
The PfEMP1s from different infected red blood cells bind to each other, causing the cells to form a bundle called a rosette that sticks to the walls of the blood vessels. This stops them from passing through the blood to the spleen, where the immune system would destroy the infected cells and the parasites within.
Attached to the blood vessels, the Plasmodium parasite continues its life cycle uninterrupted. Unfortunately for the patient, this can block blood flow in smaller vessels, causing tissue damage, inflammation and, in extreme cases, lead to organ failure.
PfEMP1s are encoded by a family of 60 var genes that are constantly changing through a process called recombination.
What do we know about PfEMP1s?
Another way the parasite escapes attack is by producing so many different PfEMP1s that the immune system can’t keep up.
PfEMP1s are encoded by a family of roughly 60 genes called var genes. Var genes are interesting because they are constantly undergoing a process called recombination – meaning they frequently shuffle genetic information between each other to create new variations of var gene and consequently, lots of new variations of PfEMP1.
To understand more about this recombination, scientists at the Wellcome Sanger Institute grew Plasmodium in human red blood cells for over a year. They’d frequently sequence the entire parasite genome to understand how it changed over different time periods. Effectively, they watched the parasites evolving in real-time.
The scientists found that var gene recombination happens in around 0.2% of parasites, every 48 hours. This might sound small – but since the average malaria patient has a billion parasites inside them, that’s roughly 2 million new var gene sequences, every two days.
What does this mean for the immune system?
The immune system is good at finding unusual things in the body, like viruses and bacteria, that need to be destroyed to keep the body safe. But when it comes to the proteins produced by the Plasmodium falciparum parasite, there’s far too much variety for the immune system to be able to keep up with.
We can think of variations of PfAMP1 proteins like different coloured ‘coats’ for the parasite. It might start off wearing a blue coat, which the immune system can recognise as dangerous – and begins to produce antibodies to make sure any blue coats are destroyed. But then the parasite switches to an orange coat, while the immune system is still looking for blue coats. By the time the immune system has learned the orange coat also needs to be attacked, the parasite has changed to a different colour coat.
The scientists worked out that through var gene recombination, the Plasmodium falciparum can create up to 60 different PfAMP1s – or coat colours – with each infection. This makes it incredibly difficult for the immune system to pin the parasite down and eliminate it from the body.

“Now, here, you see, it takes all the running you can do, to keep in the same place.”
The Red Queen in Through the Looking-Glass, Lewis Carol
The Red Queen battle
The exhausting evolutionary race of the parasite’s var recombination to produce new PfAMP1s, followed by the immune system developing new antibodies to recognise the new PfAMP1 and destroy the parasite, is sometimes called a Red Queen battle.
This is a theory based on the Red Queen’s race in Lewis Carols’ book Through the Looking-Glass. In the book, the Red Queen and Alice are running but appearing to remain in the same spot. The Red Queen comments, “Now, here, you see, it takes all the running you can do, to keep in the same place”.
In biology, it’s often used to illustrate that organisms must constantly adapt and evolve to survive in an ever-changing environment. In the case of malaria, the immune system is constantly trying to destroy the parasite to eliminate the infection. But the parasite is consistently managing to evade it by generating diversity through processes such as genetic recombination.
Add in the fact that PfEMP1 proteins are only one out of many hundreds of antigens expressed by the malaria parasite, and you can get an idea of how complicated the interaction between host and parasite becomes.
What’s next for genomics in malaria research?
Scientists continue to research how proteins like PfAMP1 help malaria parasites to survive and evolve. A deeper understanding of the genomics of malaria may open up new avenues for treatments, for example by interfering with the process of recombination to slow the parasite’s evolution.